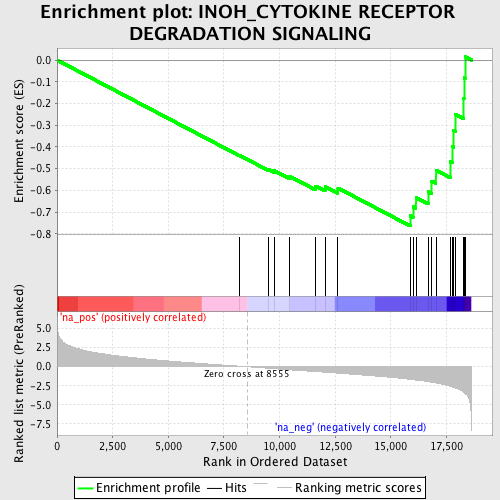
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | INOH_CYTOKINE RECEPTOR DEGRADATION SIGNALING |
| Enrichment Score (ES) | -0.7638833 |
| Normalized Enrichment Score (NES) | -2.2820435 |
| Nominal p-value | 0.0 |
| FDR q-value | 5.8319984E-4 |
| FWER p-Value | 0.0050 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMA4 | 8185 | 0.076 | -0.4379 | No | ||
| 2 | PSMB9 | 9493 | -0.199 | -0.5026 | No | ||
| 3 | PSMA5 | 9751 | -0.255 | -0.5091 | No | ||
| 4 | PSMB8 | 10463 | -0.406 | -0.5358 | No | ||
| 5 | SOCS3 | 11623 | -0.637 | -0.5800 | No | ||
| 6 | PSMB10 | 12046 | -0.713 | -0.5823 | No | ||
| 7 | PSMB7 | 12623 | -0.842 | -0.5893 | No | ||
| 8 | PSMB2 | 15870 | -1.651 | -0.7169 | Yes | ||
| 9 | PSMA6 | 15999 | -1.696 | -0.6754 | Yes | ||
| 10 | PSMC1 | 16131 | -1.742 | -0.6329 | Yes | ||
| 11 | PSMA1 | 16678 | -1.954 | -0.6066 | Yes | ||
| 12 | PSMA3 | 16833 | -2.024 | -0.5572 | Yes | ||
| 13 | PSMA2 | 17030 | -2.122 | -0.5073 | Yes | ||
| 14 | PSMB5 | 17668 | -2.567 | -0.4685 | Yes | ||
| 15 | PSMB1 | 17769 | -2.650 | -0.3984 | Yes | ||
| 16 | PSMA7 | 17818 | -2.692 | -0.3243 | Yes | ||
| 17 | PSMB4 | 17903 | -2.770 | -0.2499 | Yes | ||
| 18 | PSMD2 | 18267 | -3.295 | -0.1755 | Yes | ||
| 19 | PSMB6 | 18293 | -3.344 | -0.0816 | Yes | ||
| 20 | PSMB3 | 18339 | -3.474 | 0.0149 | Yes |